You are here
Synthetic and Systems Biology of Microalgae
Our species and civilization may be facing the most important and unprecedented agricultural, environmental, industrial and economic challenges in their history. Synthetic and systems biology offer new opportunities to answer fundamental questions using new approaches and concepts or to create artificial systems with biotechnological and industrial applications. We believe that this is a wonderful opportunity to enable a second green revolution to address the most pressing challenges by taking advantage of phototrophic microorganisms such as green microalgae and cyanobacteria, and their inherent ability to fix CO2 to produce organic molecules in a sustainable and sunlight-dependent process. Addressing these grand challenges will require a better understanding of the mechanisms of photosynthetic carbon fixation, the processes that enable adaptation to environmental stresses, and the redox signaling network that tightly controls both processes. The team is interested in these three themes and is also developing methods, tools and concepts of synthetic biology in microalgae for fundamental and applied developments, as well as technologies for digital storage on DNA.
The team uses multidisciplinary approaches combining systems biology, synthetic biology, molecular biology, biochemistry, chemobiology, protein engineering, proteomics, structural biology, genetics and cell biology.
Our studies are mainly focused on the unicellular green alga Chlamydomonas reinhardtii, a major model organism for the study of photosynthesis and having a strong biotechnological potential. Complementary models are also studied such as the yeast Saccharomyces cerevisiae, the cyanobacterium Synechocystis PCC6803, the terrestrial plant Arabidopsis thaliana or the bacteria Escherichia coli.
The work of our team focuses on five research areas described below
DNA Data StorageProject Leader: The storage of digital data is a major challenge in our modern societies. Current digital media stored in data centers are fragile, bulky and energy-intensive. The ongoing digital transformation of our societies requires a major disruption of our digital storage technologies. DNA data storage is an emerging technology that has the advantage of being stable for thousands of years, extremely compact and energy-efficient. Our team is developing innovative concepts of DNA data storage based on domestication of biological systems  |
Synthetic Biology of MicroalgaeProject Leader: Synthetic biology is the forward engineering of biological systems using an ever-expanding list of biological functional modules. It aims at understanding by building biological systems through rational design. We use its concepts and tools to study photosynthesis in the model green alga Chlamydomonas reinhardtii to better understand the regulation and limitations of photosynthetic carbon fixation. Our first approach is the redesign of pathways to control in vivo the amount of each enzyme. The second project (paleo-biochemistry) aims at using Evolution as a resource (ancestral sequences inference and resurrection, directed evolution) to explore novel evolutionary paths of carbon fixation. We also aim at implementing in vivo new metabolic pathways (de novo design) to expand photosynthetic capacities. Finally, we develop the standardized tools (MoClo, logic gates) necessary to build these designs.  |
Structure and Interactions of Calvin-Benson EnzymesProject Leader: The experimental determination of the structure of the twelve proteins of the Calvin-Benson pathway of photosynthetic carbon fixation revealed the biochemical mechanisms by which enzymes catalyze these essential reactions for the biosphere. We more specifically study how post-translational modifications (oxido-reductions and phosphorylations) modulate enzyme activities, and to which extent protein-protein interactions connect enzymes together or with their thioredoxin regulators. The full description of allosteric mechanisms and channeling shortcuts of the Calvin-Benson system is further explored by the design-building-testing of artificial multiprotein complexes.  Photosynthesis is the main energy and carbon source for the biosphere. The Calvin-Benson cycle fixes atmospheric CO2 into organic trioses in the stroma of chloroplasts. While the 11 enzymes individually catalyzing the chemical steps of the cycle are known, little has been reported about their supra-molecular organization. Hence, the CALVINTERACT project ambitions to redefine the molecular basis for Calvin-Benson photosynthetic carbon fixation, in the light of the regulatory interactions that emerged between its protein components. We propose to determine the crystal structures of three multi-protein complexes, to measure their protein-protein interactions in vitro and to detect native complexes from cell extracts. The study will be completed with the design of artificial interactions in a synthetic biochemistry perspective. Physiological effects will be assayed on the model unicellular alga Chlamydomonas reinhardtii.
The project ambitions to define in a model unicellular organism, the green alga Chlamydomonas reinhardtii, the structures of all Calvin-Benson enzymes, a pathway that ensures biological carbon fixation. Besides the determination of novel individual structures for chloroplastic enzymes, the project aims at describing homo-oligomerization of these proteins, their interactions with thioredoxin redox regulators, and protein-protein interactions that enzymes establish together. In a synthetic biology approach, artificial interactions are designed and their effects tested on enzymatic efficiency and metabolism.
The project mainly depends on the determination of high-resolution structures by X ray crystallography, with recombinant proteins overexpressed in the bacterial host Escherichia coli. During the first months of the project, we submitted one of the enzymes to electron microscopy analysis. Complexed states of Calvin Benson enzymes are searched for in algae extracts, by chromatographic means to begin with, then by co-immunoprecipitation in the future. An in-silico strategy of protein-protein interaction prediction was set up recently in collaboration with experts in computational biology.
Structures of all remaining enzymes, except one, were obtained. A novel redox regulator was revealed by the identification of surface residues in the crystallographic model. High molecular weight species of several enzymes were observed from size-exclusion chromatography, justifying further validation and characterization. The in vitro isolation of multi-enzymatic complexes and the de novo design of multi-protein assemblies are the prominent objectives of the second phase of the project.  |
Redox Signaling NetworkProject Leader: Responses to fluctuating environmental conditions are based on sophisticated signaling and metabolic regulation networks where reactive oxygen, nitrogen and sulfur species play a key role. They act mainly on specific protein cysteines by a set of redox post-translational modifications (disulfide bridges, nitrosylation, glutathionylation, persulfidation...). These modifications are controlled by the small oxidoreductases thioredoxins and glutaredoxins. We are developing, mainly in Chlamydomonas reinhardtii, innovative approaches combining chemobiology, synthetic biology and systems biology to 1/ understand the dynamics and plasticity of these redox modification networks and 2/ understand at the molecular level how reactive species, redox modifications and redox proteins act together to rapidly modulate cellular processes and metabolic pathways in order to allow adaptation and survival.  |
Environmental Stress ResponsesProject Leader: We use Chlamydomonas to try to understand how a unicellular organism adapts to survive in a hostile environment. If it has already been shown that in the face of a strong stress Chlamydomonas can trigger programmed cell death (PCD), we have discovered in our team that in response to more moderate stresses, cells aggregate forming impressive multicellular structures in which algae can be protected from the toxic environment. We are trying to uncover the mechanisms that control both PCD and aggregation in Chlamydomonas. 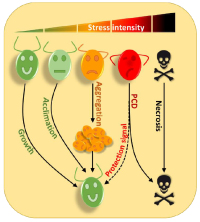 Chlamydomonas reinhardtii is the most prominent model organism in the green algae lineage for both basic research and biotechnological applications. We use Chlamydomonas to try to understand how a unicellular organism adapts to survive in a hostile environment. Although some experiments indicate the existence of programmed cell death (PCD) in response to severe stresses in Chlamydomonas, the mechanisms controlling this process and the signaling pathways involved, have yet to be identified (de Carpentier et al., 2019). Excitingly, we have shown recently that Chlamydomonas cells that self-destruct emit molecules in the culture medium that allow better resistance and survival of other cells in the population, highlighting the existence of altruistic mechanisms. We have also discovered that in response to more moderate stresses, cells aggregate forming multicellular structures in which algae can be protected. Cellular aggregation would be an intermediate stage where the stress is not strong enough to induce PCD, but where nevertheless cells collaborate to protect themselves.
As Chlamydomonas is amenable to powerful genetic approaches, we have built an insertional mutant library, from which we have identified two families of original mutants: the survivor (sur) mutants which are no longer triggering PCD in response to several stresses, and the socializer (saz) mutants that form aggregates spontaneously. We have identified the genes affected in most of these mutants. Further characterization of the mutants by RNA-Seq and quantitative proteomic studies has been performed and allowed us to have a better overview of the mechanisms controlling PCD and aggregation in Chlamydomonas. Our multi-omics approach led us to identify potential key regulators of PCD and aggregation in Chlamydomonas. These candidates are then tested for their involvement in the aggregation or PCD using mutants obtained from the Chlamydomonas Library Project (CLiP) (Li et al., 2019).
Since the dynamics and mode of formation of multicellular structures in Chlamydomonas are very poorly known, we are also developing microfluidic tools that allow us to follow this process in detail over a period of time ranging from several hours to several days. Some of the candidates we have identified in our multi-omics studies can be fused to specific fluorescent proteins, using tools developed in our lab (Crozet et al., 2018; de Carpentier et al., 2020), to follow their movement during the same period, and to highlight their potential role.
The aggregation of cells over a long period of time is one of the hypotheses classically formulated to explain the transition from unicellularity to multicellularity over the course of evolution. Our research on the mechanisms leading to aggregation in response to stress and the characterization of saz mutants could therefore also contribute to a better understanding of the appearance of multicellularity in Volvocales. References:  |




